
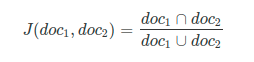

Where # is determined using rarefaction.Code: * Example generated by -dataex. Qiime diversity core-metrics-phylogenetic -i-phylogeny rooted-tree.qza -i-table bacteria-table.qza -p-sampling-depth # -m-metadata-file mdat.tsv -output-dir core-metrics-results I do run the following command to get some core metrics: Where State is the categorical variable chosen from the metadata file. Qiime diversity beta-group-significance -i-distance-matrix jaccard_table.qza -m-metadata-file mdat.tsv -m-metadata-column State -o-visualization jaccard_table.qzv Qiime diversity beta -i-table bacteria-table.qza -p-metric jaccard -o-distance-matrix jaccard_table Qiime diversity alpha-group-significance -i-alpha-diversity shannon_table.qza -m-metadata-file mdat.tsv -o-visualization shannon_table.qzvįor Jaccard’s similarity I ran the following: Qiime diversity alpha -i-table bacteria-table.qza -p-metric shannon -o-alpha-diversity shannon_table Thus, it appears that all of my samples are included in the analyses. Okay, one last question (I think)- is there a way to integrate sampling depth into these analyses? As I currently have the arguments structured, they are using a table.qza (specifically bacteria-table.qza, as specified in my earlier post) that hasn’t accounted for sampling depth.


 0 kommentar(er)
0 kommentar(er)
